All photos by Dr. Pete Countway
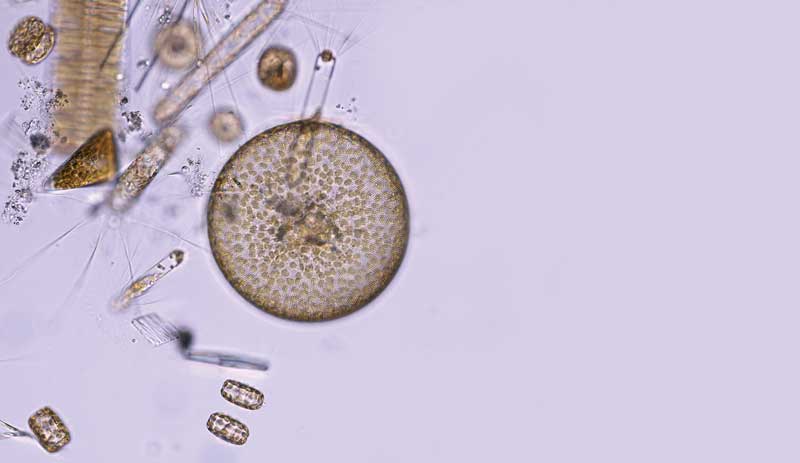 An array of the near-invisible life that drifts in the ocean surface. This micrograph was taken in the waters off Antarctica.
An array of the near-invisible life that drifts in the ocean surface. This micrograph was taken in the waters off Antarctica.
Techniques for observing the ocean have come a long way over the centuries. Our early pictures come from anecdotes from people like Theophrastus, a student of Aristotle’s, who observed, “If many jellyfish appear in the sea, this is a sign of a stormy season.” Some 2,000 years later, Benjamin Franklin, with information from his whaling cousin, made the first maps of the “Gulph Stream.”
As scientific tools have advanced, our picture of the ocean has become more and more detailed. The launching of satellites, for example, was like turning on a light in a dark room: suddenly the huge monotonous ocean was lit up with spiraling multi-colored swirls of currents and plankton. With each technological advance, we see more details—knowledge that is crucial to understanding how the ocean makes life on Earth possible.
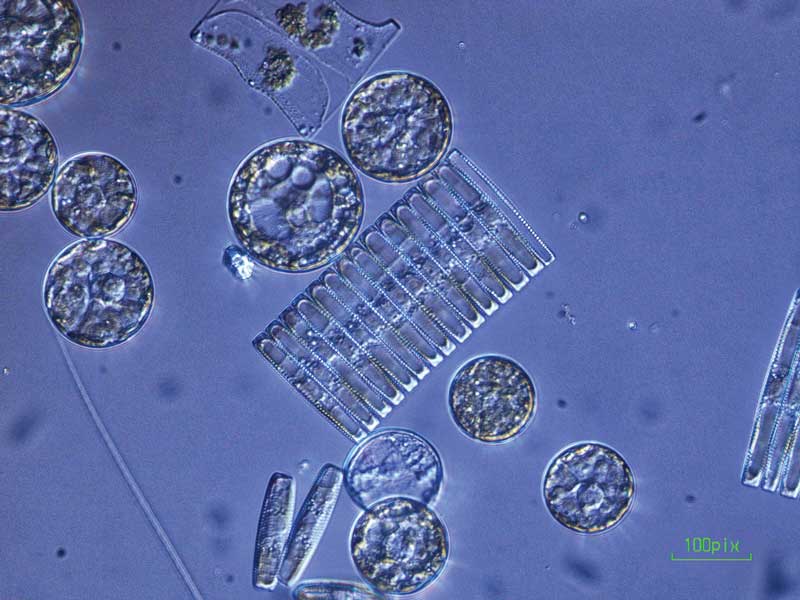 Plankton come in a beautiful variety of shapes and sizes. But often, the best way to distinguish species—for example telling a toxic species from a nontoxic one—is by inspecting its DNA.
Plankton come in a beautiful variety of shapes and sizes. But often, the best way to distinguish species—for example telling a toxic species from a nontoxic one—is by inspecting its DNA.
Much of the ocean’s staggering diversity of life has been hidden in dark, impenetrable depths. Send down a probe—or better yet, travel down yourself in a submersible—and you will struggle to see beyond a short radius. Yet the life within even a single drop of seawater contains far more genetic diversity than all of humanity. Imagine this diversity spread across the swirling currents, the deep ocean trenches and hydrothermal vents, from the tropical seas to the icy poles. This complexity of life has been difficult and expensive to observe and map, but a new revolution in ocean science could change that.
It involves environmental DNA, or eDNA for short.
Let’s start with a simple question: What is a fish? Scientifically speaking, it’s a cold-blooded aquatic vertebrate, whose persona can be perfectly mimicked by squeezing one’s cheeks together. A more complex answer comes from Mike Kinneson, a biologist at the University of Maine, who once described a fish as “a leaky bag of genes that does something in water.” In other words, a fish is a swimming sack of DNA. That DNA is the key to this revolution in ocean science.
The next time you’re at the seaside, dip a bottle in the ocean, and hold it up to the light. It might be clear, bubbly, or even a little cloudy. If you’re lucky, you might see some tiny speck swimming around inside. But the amazing thing about this bottle of seawater is what you can’t see, which is probably the most complex and powerful potion known to humankind. If you were able to zoom in by a factor of roughly one zillion, and examine all of the DNA in that bottle, a vast universe would be revealed. If you zoom in far enough, you might even see a whale.
 A smartphone connected to the BioMeme Two3, a device that can be used to detect and identify DNA in the environment. Photo by Dr. Pete CountwayStart by zooming in to the power of ten. This would reveal the larvae of fish, mussels, oysters, starfish, and other denizens of the deep. Zoom in farther and you might see tiny flea-like crustaceans, like copepods, and large single-celled algae. These latter produce much of the oxygen that we breathe, and also absorb carbon dioxide. Keep zooming in and you would see billions of bacteria and archaea, and then marine viruses, which, taken en masse, are so abundant as to possibly outweigh all other forms of life in the ocean.
A smartphone connected to the BioMeme Two3, a device that can be used to detect and identify DNA in the environment. Photo by Dr. Pete CountwayStart by zooming in to the power of ten. This would reveal the larvae of fish, mussels, oysters, starfish, and other denizens of the deep. Zoom in farther and you might see tiny flea-like crustaceans, like copepods, and large single-celled algae. These latter produce much of the oxygen that we breathe, and also absorb carbon dioxide. Keep zooming in and you would see billions of bacteria and archaea, and then marine viruses, which, taken en masse, are so abundant as to possibly outweigh all other forms of life in the ocean.
And we’re not done yet.
You’ll have to zoom in again to see the whale I mentioned. This all is because the seawater in your bottle is filled with tiny bits of DNA shed from all kinds of living things—a.k.a. the “leaky bags of genes”—from bacteria to whales. Anything that has passed through these waters recently has left its DNA signature. Think of it as looking at the ocean and seeing the trails of fish, jellies, sea turtles, octopuses, and even whales.
The technology to read DNA has been around for a few decades, getting its start in forensics. In 1985, in Leicestershire, England, more than 5,000 men lined up to give blood and saliva samples for DNA profiling. At the time, DNA profiling was a brand new sort of wizardry with the potential to identify the killer of two young Leicestershire girls. This was a new concept for criminal investigations—the idea that everywhere you go, you leave tiny bits of yourself that can be detected with DNA technology. Somewhat ironically, the killer was caught, not by the DNA profiling, but rather the mere idea of it. As folks were lining up for the test, the killer, afraid to be caught, sent someone else to take the test in his place. Old-fashioned detective work uncovered his deception and eventually led to his confession, without him actually ever taking the DNA test. Nevertheless, the door had been opened, and DNA technology is now a common tool in crime investigations.
The same transition is happening today for eDNA. A few months ago, if you typed eDNA into Google, you would get a list of information about people named Edna. Now, the top hit is the quickly expanding Wikipedia page for environmental DNA. Today, it’s possible to dip a bottle in the ocean, fill it with seawater, and then “interrogate” it for all of the tiny traces of DNA floating by.
As animals move to and fro across the ocean, they constantly shed molecules, leaving DNA-shaped paths behind them. That means a bottle of seawater can tell us if any protected salmon or endangered whales are in the neighborhood, or if an endangered sea turtle or the enigmatic giant squid has passed through. It can tell us if we’re in the vicinity of pathogens or toxic red tides invisible to the unaided eye. And if we dig deep enough into the DNA, we can ask questions about how life originated and evolved.
One research group has even tested the waters of Loch Ness for signs of dinosaur DNA, hoping to finally demonstrate the existence of Nessie; the closest thing they found was the DNA signal of eels.
It’s not hard to imagine how helpful this new technology is for ocean science. By dipping bottles into the ocean and peering inside at the DNA, we can pull back the thick curtain that obscures life under the waves. Even more exciting is the way in which eDNA might democratize science. Over the past 10 years or so, the cost of sequencing DNA has dropped drastically.
The technology has reached the point where soon pretty much everyone will be able to use it. Student groups have already used DNA technologies to uncover that something labeled “shark fin soup” was actually made of skate, and to reveal that sometimes fish for sale in the market are not the species listed on the label.
In the classic TV show Star Trek, space explorers had a small rectangular device, about the size of a smartphone, called a “tricorder.” They could hold it up like a wand and scan the environment or the human body. Amazingly, the smartphone itself could soon be a working tricorder. A company called BioMeme has designed a small device called the Two3 that hooks up to a smartphone.
Using the Two3, you could take water from any environment (ocean, stream, or even the human body), and scan it for signs of DNA. The Two3 and other similar devices are becoming more accessible, putting the ability to read DNA into the hands of anyone with a smartphone. This opens the door to many applications. At Bigelow Laboratory in East Boothbay, for example, scientists are working with shellfish growers and managers to use the technology to monitor for toxic red tides in real time. The device is simple enough that it can be used at the dock, requiring only a bottle of seawater. Others are hoping to equip mariners with the ability to check waters for evidence of endangered species like right whales. The data comes up instantly, without any need to send samples off to a lab. As more and more users of the technology make measurements in the ocean, we’ll be able to fill in the map with the diverse mosaic of life under the waves.
Putting technologies like these in the hands of everyday people is becoming more important. As the climate changes faster and faster, the frequency and intensity of environmental hazards is growing. From toxins in drinking water to resource collapses, it’s getting harder for public agencies to stay on top of these hazards. This also comes at a time when many of the same agencies are deliberately relaxing their oversight and regulations, leaving us all vulnerable. Enabling everyday people to fill a bottle with seawater and read the eDNA around us will better equip us to foresee and cope with some of the hazards coming our way.
Nicholas R. Record, PhD is a senior research scientist at Bigelow Laboratory for Ocean Sciences. Disclosure: He shares a distant ancestor with all modern-day DNA-carrying organisms.
Maine lands major grant for DNA research
Maine is on the cutting edge of using DNA to gain a better understanding of coastal ecosystems. Last summer, the University of Maine and the Bigelow Laboratory for Ocean Sciences announced that they had been awarded a $20 million grant from the National Science Foundation EPSCoR program for a five-year initiative that aims to use new eDNA technology to revolutionize environmental monitoring, ecological understanding, and sustainability of coastal ecosystems. The two lead organizations will work on the project with other collaborators in education, government agencies, citizen’s groups, and local industry statewide.
The new program will focus on two pressing issues for the coast of Maine: sustainable fisheries and harmful species. Sustainable fisheries research includes studying the outcomes of large-scale restoration efforts and unraveling the complex early life cycles of economically important species like lobster and scallops. Work on harmful species includes developing early warning systems for toxic algal blooms, and forecasting the spread and impacts of invasive species.
The program also will advance the next generation of environmental science by combining the genetic data across these projects, and other sampling efforts, to generate massive genetic databases that researchers, policymakers, and industry members can access to address coastwide challenges for decades to come.
Citizen scientists, from seafood growers to schoolchildren, will have opportunities to contribute in multiple ways, such as collecting water samples, monitoring local water sources with handheld eDNA detectors, and providing vital input for future initiatives.
How cool is that!?
—The Editors






